Dynamics of microbial communities
Investigation of the mixed culture’s dynamic in batch and chemostat culture (Christian Riedele):
With a recently advanced t-RFLP method, it is possible to quantify the cell number of a single bacterial species in a mixture of several different bacteria. We are using this method to study the behaviour of the 3 species mentioned above in bioreactor time-series experiments on the level of species specific cell counts and extracellular metabolites. In combination with mathematical modelling we focus on finding interactions between the bacteria in mixed culture in comparison to the pure cultures’ dynamics. Currently, we are investigating the mixed culture’s reaction onto antibiotic pulses in so called “time-kill” experiments.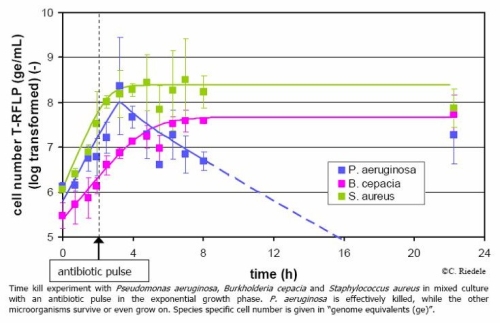
Assessment of the state of single cells by flow cytometry (Marc Rüger)
Flow cytometry allows for multiparametric representation of individual cells within a population [1]. Hence, heterogeneity of a population can be assessed on a single cell level. Therefore it has been applied to the bacterial consortium of interest.
In our studies we are particularly interested in the assessment of viability of each species within this mixed community. This may allow for a more detailed description of mixed culture dynamics and may provide new insights into interspecies interaction. In order to detect viability on a species level, species-specific immunofluorescent probes and Gram-specific fluorescence probes are applied. Viability is determined by membrane integrity analysis using dead cell marker propidium iodide in combination with total cell staining by SYBR®Green I.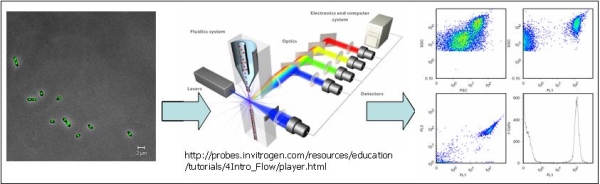
Workflow of flow cytometric analysis: 1.) Staining: Fluorescence microscope picture of Burkholderia cepacia cells stained against DNA with SYBRgreenI (LSM 510, Zeiss, 1000 x); 2.) Analysis: Principle of multiparametric flow cytometry; 3.) Data analysis: Representation of Scatter and Fluorescence data of SYBRgreenI stained Burkholderia cepacia cells in histograms and 2 D-plots, respectively.
[1] Davey H.M. et al. (2003). Current Issues in Molecular Biology 5: 9 - 15
Describing interactions on molecular level by metaproteome analysis (Dirk Benndorf, Sabine Kluge, Erdmann Rapp)
For improving our models we want to describe the interactions on the molecular level by metaproteome analysis. Currently, proteins extracted from mixed cultures can be separated by 2D-PAGE. Based on pattern comparison and identification by mass spectrometry, many protein spots can be assigned exclusively to one species. The quantification of house-keeping proteins can be used for calculating the proportion of each species in the mixed culture. Further experiments using this approach will be carried out to analyze the effects of cultivations conditions or addition of antibiotics on interactions of B. cepacia, P. aeruginosa and S. aureus in our model cultures.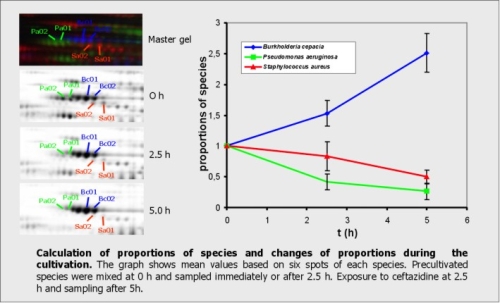
References
[2] Hesseler J, Schmidt JK, Reichl U, Flockerzi D (2006). Coexistence in the chemostat as a result of metabolic by-products. J Math Biol. 53, 556-584.
[3] Schmidt, J. K., König, B., Reichl, U. (2007). Characterization of a three bacteria mixed culture in a chemostat: evaluation and application of a quantitative terminal-restriction fragment length polymorphism (T-RFLP) analysis for absolute and species specific cell enumeration. Biotechnol. Bioeng. 96, 738-756.
[4] Schmidt JK, Riedele C, Regestein L, Rausenberger J, Reichl U (2011). A novel concept combining experimental and mathematical analysis for the identification of unknown interspecies effects in a mixed culture. Biotechnology and Bioengineering 2011, 108(8), 1900-1911
[5] Riedele C, Reichl U (2011). Interspecies effects in a ceftazidime-treated mixed culture of Pseudomonas aeruginosa, Burkholderia cepacia and Staphylococcus aureus: analysis at the single-species level. Journal of Antimicrobial Chemotherapy 2011, 66(1), 138-145
[6] Kluge, S,Hoffmann, M., Benndorf, D., Rapp, E., Reichl, U. (2012). Proteome Based Tracking and Analysis of a Bacterial Mixed Culture. Proteomics 12, 1893-1901. (PubMed).
[7] Riedele C, Reichl U (2012) Time-kill studies with a ceftazidime-treated mixed culture consisting of Pseudomonas aeruginosa, Burkholderia cepacia and Staphylococcus aureus. Engineering in Life Sciences 2012, 12 (2), 188-197
[8] Rüger M., Bensch G., Tüngler R., Reichl U. (2012) A Flow Cytometric Method for Viability Assessment of Staphylococcus aureus and Burkholderia cepacia in Mixed Culture. Cytometry Part A, Volume 81A, Issue 12, 1055 1066 (Pubmed)
[9]Rüger, M., Ackermann, M., & Reichl, U. (2014). Species-specific viability analysis of Pseudomonas aeruginosa, Burkholderia cepacia and Staphylococcus aureus in mixed culture by flow cytometry. BMC Microbiology, 14, 56. (PubMed)




