Metaproteomics of Microbial Communities in Biogas Plants
Responsible: Dirk Benndorf
Contributors: Robert Heyer, Kay Schallert (PhD student), Patrick Hellwig (PhD student), Sebastian Püttker (PhD student)
More than 9.000 biogas plants (BGP) in Germany produce about 5% of the renewable electricity. Biogas yields and process efficiency depend on the metabolic abilities and performances of the single microorganisms. Design, optimization and control of biogas processes is challenging since the impact of microbial communities on biogas production as well as on process stability and efficiency is poorly understood.
A workflow for metaproteomics of biogas plants was developed and applied frequently to characterize the structure of microbial communities and infer about their metabolic performance in biogas digesters.
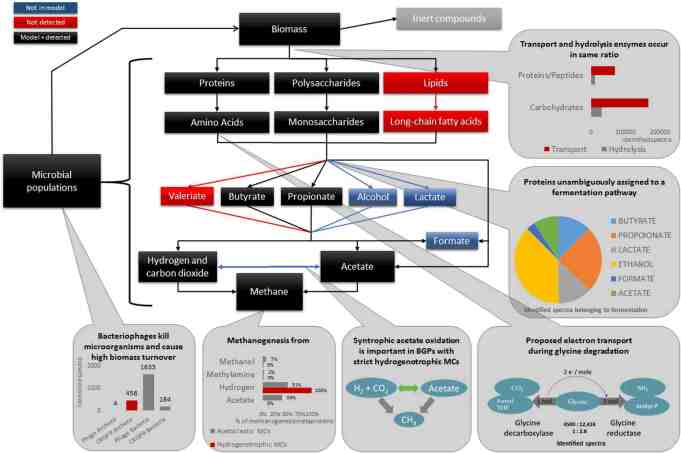
Microbial communities of biogas plants could be partly related to functional properties and to different process conditions. For all biogas plants examined, two main types of microbial communities were observed. The first type comprised communities, which performed acetoclastic and hydrogenotrophic methanogenesis simultaneously. In contrast, the second type showed strictly hydrogenotrophic behaviour but the presence of large amounts of proteins of the bacterial C1-metabolism implicated syntrophic acetate oxidation. Furthermore, metaproteomic studies suggests that competition and host-phage interactions affect the taxonomic and functional composition of microbial communities.
Figure 1: Mapping of the identified metaproteins to the Anaerobic Digestion Model 1 (ADM1). Identified metaproteins were assigned to individual steps of the ADM1. Significant differences between the assumed steps in the ADM1 and steps confirmed by the identified metaproteins are highlighted in RED or BLUE. Aspects not covered by metaproteomics analysis are displayed in GRAY (e.g., “Inert compounds”). For each of the analyzed steps a summary provides the most important findings of this study. MCs: microbial communities. Figure taken from Heyer et. al (2019) [8].
Collaborations:
- Susanne Theuerl, Katharina Willenbücher (Leibniz-Institut für Agrartechnik und Bioökonomie)
- Andreas Schlüter, Alexander Sczyrba (CeBiTec, Universität Bielefeld)
Funding:
- Deutsche Bundesstiftung Umwelt e.V. (DBU), PhD grant for Robert Heyer (01.08.2011-31.07.2014): Prozesskontrolle und Optimierung der Biogasproduktion mittels Metaproteomanalyse.
- BMELV, FNR, FKZ 22028811, Cooperative project (01.11.2011-31.10.2013): Determination of the microbial diversity present in biogas plants and of technological factors with major impact on the microbial community structure (BIOGAS-BIOCOENOSIS), workpackage: Metaproteome analysis for monitoring hydrolytic and methanogenic activities of microbial communities in biogas plants analysis.
- BMELV, FNR, FKZ 22404115, Cooperative project (01.12.2015-30.11.2019): Biogas-Monitoring III, project 2: systems microbiology, workpackage 3: enzymatical biodiversity.
- BMEL, FNR, FKZ 22403816, Cooperative project (01.06.2017-31.05.2020):biocatalysts in bioreactors: monitoring, control and und optimization of biogas production, workpackage: measurement of enzyme and protein composition.
References:
- Hanreich, A., Schimpf, U., Zakrzewski, M., Schlüter, A., Pühler, A., Benndorf, D., Rapp, E. Reichl, U., Heyer, R., Klocke, M. (2013). Metagenome and metaproteome analyses of microbial communities in mesophilic biogas-producing anaerobic batch fermentations indicate concerted plant carbohydrate degradation. Systematic and Applied Microbiology 36, 330-338. PubMed
- Heyer, R., Kohrs, F. , Benndorf, D., Rapp, E. , Kausmann, R. , Heiermann, M. , Klocke, M., Reichl, U. (2013). Metaproteome analysis of the microbial communities in agricultural biogas plants. New Biotechnology 30, 614-622. PubMed
- Kohrs, F., Heyer, R., Magnussen, A., Benndorf, D., Muth, T., Behne, A., Rapp, E., Kausmann, R., Heiermann, M., Klocke, M., Reichl, U. (2014). Sample prefractionation with liquid isoelectric focusing enables in depth microbial metaproteome analysis of mesophilic and thermophilic biogas plants. Anaerobe 29, 59-67. PubMed
- Theuerl, S., Kohrs, F., Benndorf, D., Maus, I., Wibberg, D., Schlüter, A., Kausmann, R., Heiermann, M., Rapp, E., Reichl, U., Pühler, A., Klocke, M. (2015). Community shifts in a well-operating agricultural biogas plant: how process variations are handled by the microbiome. Appl Microbiol Biotechnol., 99, 7791-7803. PubMed.
- Heyer, R., Kohrs, F., Reichl, U., Benndorf, D. (2015). Metaproteomics of complex microbial communities in biogas plants. Microb. Biotechnol., 8, 749763. PubMed.
- Heyer, R., Benndorf, D., Kohrs, F., De Vrieze, J., Boon, N., Hoffmann, M., Rapp, E.,Schlüter, A., Sczyrba, A., Reichl, U. (2016) Proteotyping of biogas plant microbiomes separates biogas plants according to process temperature and reactor type. Biotechnol Biofuels 9,155. PubMed
- Heyer, R., Schallert, K., Büdel. A., Zoun, R., Dorl, S., Behne, A, Kohrs, F., Püttker, S., Siewert, C., Muth, T., Saake, G, Reichl, U., Benndorf, D. (2019). A robust and universal metaproteomics workflow for research studies and routine diagnostics within 24 h using phenol extraction, FASP digest, and the MetaProteomeAnalyzer. Front. Microbiol., fmicb.2019.01883. Pubmed
- Heyer, R., Schallert, K., Siewert, C., Kohrs, F., Greve, J., Maus, I., Klang, J., Klocke, M., Heiermann, M., Hoffmann, M., Püttker, S., Calusinska, M., Zoun, R., Saake, G., Benndorf, D., Reichl, U. (2019). Metaproteome analysis reveals that syntrophy, competition, and phage-host interaction shape microbial communities in biogas plants. Microbiome 7, 69. Pubmed