Biomolecular analysis of dynamic interactions between influenza viruses and host cells
For a better understanding of influenza virus replication dynamics in vaccine production using adherent mammalian MDCK cells, a structured mathematical model has been developed [1]. However, detailed modeling of gene-specific transcription and replication for each class of intracellular viral RNA was only partially possible due to a lack of experimental data. Therefore, a quantitative realtime-PCR assay was developed to analyze transcription and replication of viral gene segments. The discrimination between replication (vRNA (-), cRNA (+)) and transcription steps (mRNA (+)) is possible by using polarity-specific and tagged primers during reverse transcription and realtime PCR. Based on this sophisticated method for the precise quantification of viral RNAs, validation and improvement of the existing model is supported. Furthermore, this systems biology approach will lead to a quantitative understanding of influenza virus replication and its regulation, and enable testing of hypothesis generated from experimental data sets.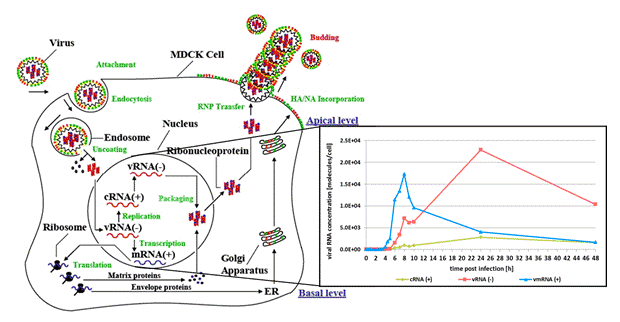
Fig 1: Hypothetical model of influenza A virus replication in MDCK cells with detection of viral RNAs of gene segment 4 (HA).
[1] Sidorenko, Y. and Reichl, U. (2004): Structured model of influenza virus replication in MDCK cells, Biotechnology and Bioengineering, 88(1), 1-14
Link to other projects of Bioprocess Engineering
Model-based investigation of influenza virus replication in mammalian cell culture




